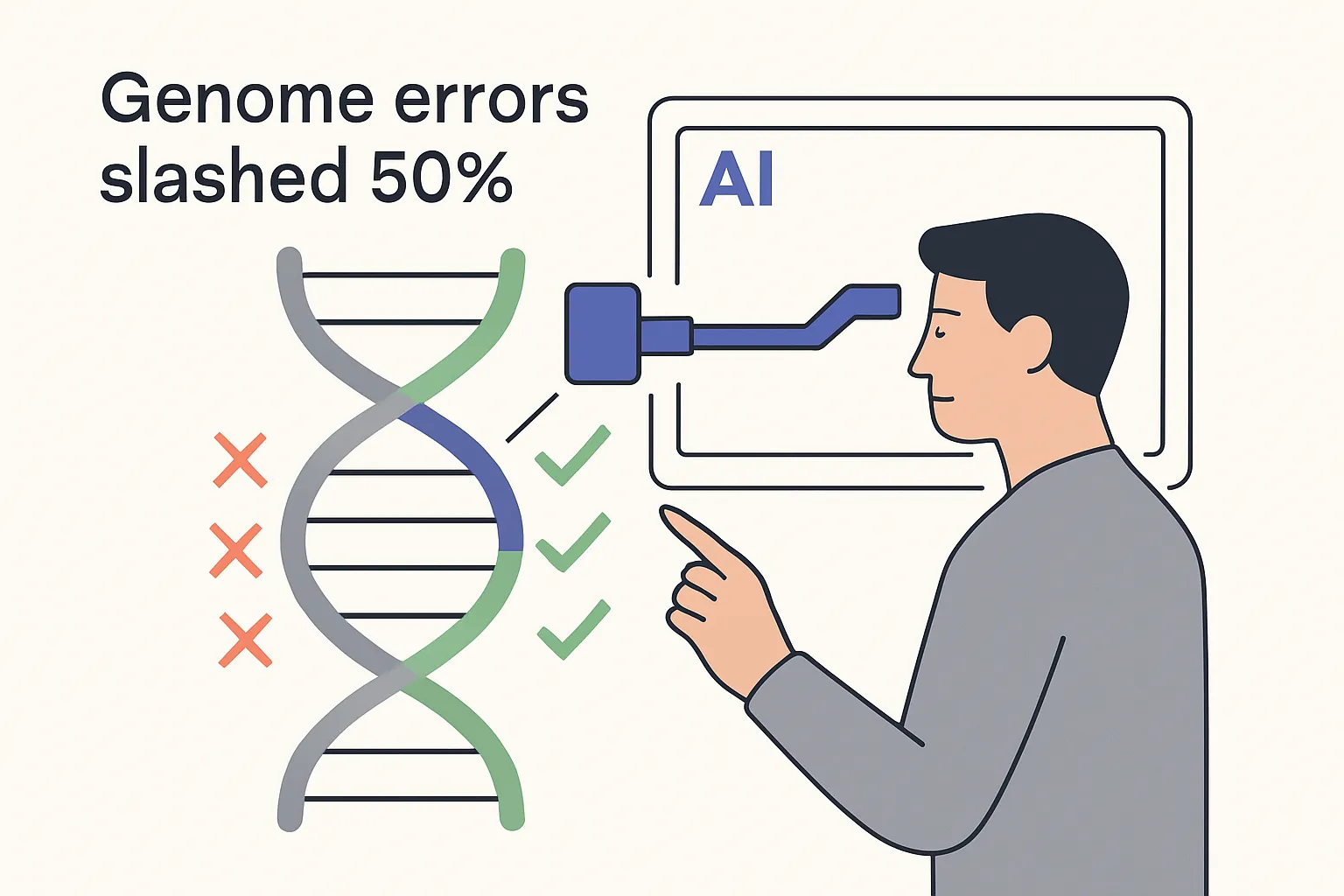
On 6 August 2025, Google Research introduced DeepPolisher, an open-source deep learning tool that reduces base-level errors in human genome assemblies by about 50 percent. Technical lead Kishwar Shafin and product lead Andrew Carroll outlined the release in a Google Research blog post from Mountain View, California.
What DeepPolisher Does
The software applies a Transformer model to correct draft assemblies generated from Pacific Biosciences long reads. The model ingests base calls, quality scores, mapping confidence and mismatch flags as separate channels, then predicts the most probable true sequence.
The project is a joint effort between Google Research and the UC Santa Cruz Genomics Institute and is released under the Apache 2.0 license.
Key Performance Results
- Total assembly errors cut in half in initial benchmarks.
- Insertion-deletion (indel) mistakes fall by 70 percent, a critical improvement because indels can shift the reading frame of protein-coding genes.
- Average assembly quality rises from Q66.7 to Q70.1 - a Q-score increase that translates to roughly 1 error in every 10 million bases.
- Training used chromosomes 1–19 of the Personal Genomes Project HG002 sample, while chromosomes 20–22 were held out for validation.
- The May 2025 production model polished 232 assemblies for the Human Pangenome Reference Consortium.
Why Genome Polishing Matters
Short-read sequencers, often developed by Illumina, achieve high accuracy but read only a few hundred bases at a time. Long-read platforms span tens of thousands of bases yet leave residual errors that complicate gene annotation and structural-variant discovery.
Earlier pipelines pushed error rates below 0.1 percent, still leaving thousands of mistakes in a single human genome. Google and PacBio previously reduced single-pass errors with DeepConsensus - the first demonstration of this on a human genome. DeepPolisher builds on that work by training on the well-characterized HG002 reference, validated by NIST and the NHGRI.
Using phased data allowed the model to learn maternal and paternal haplotype differences, improving correction accuracy across the genome.
Availability
Source code, pre-trained models and documentation are available in the public Code Repo. A peer-reviewed Genome Research paper details the architecture and benchmarking methodology.
The training sample comes from the Personal Genomes Project, providing transparent access to raw data for further study.
Further Reading
- Google Research blog announcement
- Images from NHGRI
- Background on DNA copying machinery
- Concepts behind combinations of nucleotides used in error evaluation




.svg)