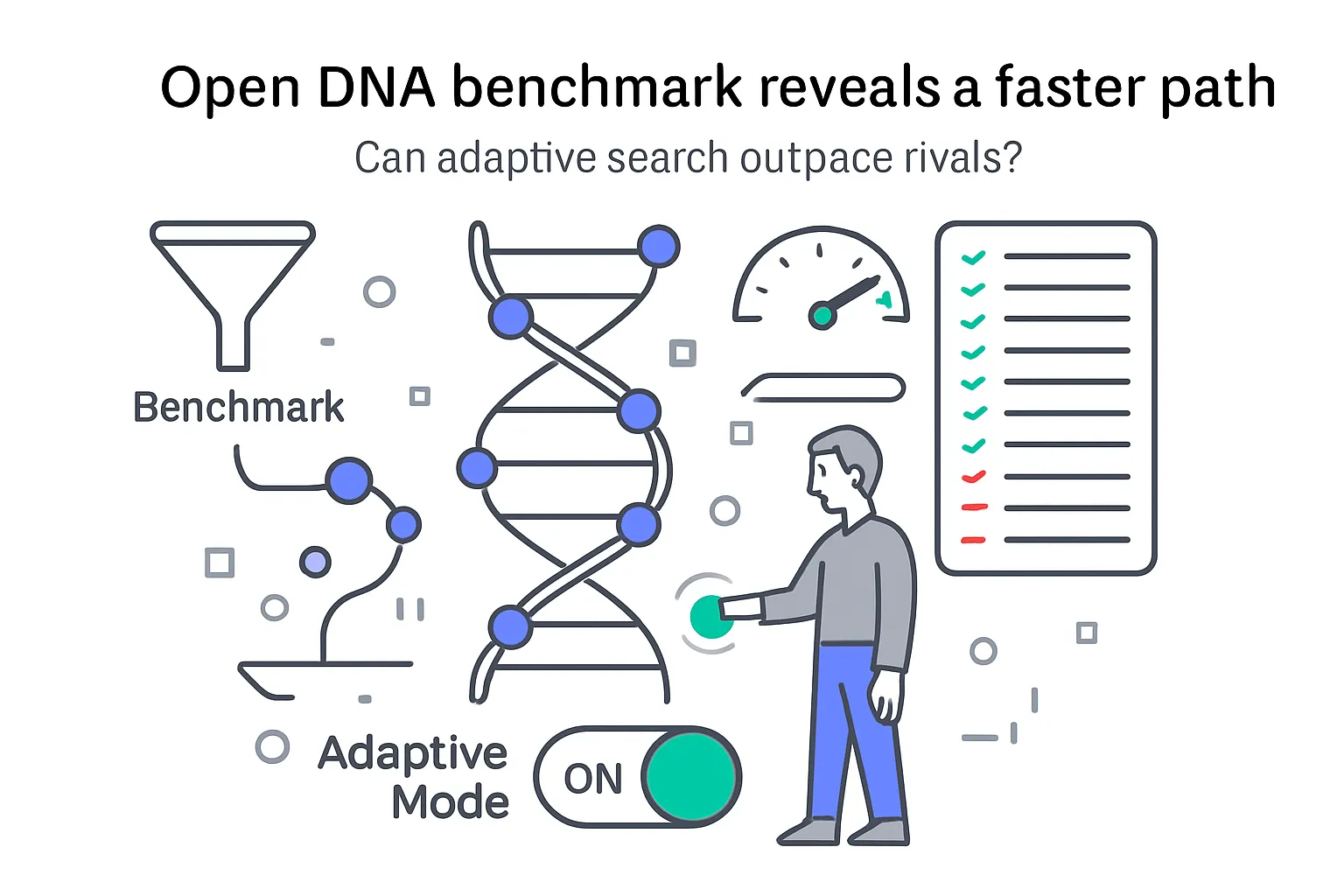
Google Research announced NucleoBench and AdaBeam on September 11, 2025, introducing an open-source benchmark for nucleic acid sequence design and a new optimization algorithm. The news, published by Cory Y. McLean on the Google Research blog, standardizes how DNA and RNA design methods are evaluated and delivers a faster, more memory-efficient search strategy that topped most tasks in the suite. Read the announcement.
What was announced
- NucleoBench - an open evaluation suite spanning 16 sequence design tasks with shared starting sequences and fixed time budgets.
- AdaBeam - a new algorithm that combines unordered beam search with adaptive strategies, designed for long genomic contexts.
- The project is a collaboration between Google Research and Move37 Labs.
- More than 400,000 experiments were run, evaluating nine algorithms under identical controls.
- Code and packages are open source: NucleoBench repository and the nucleobench Python package.
Inside the benchmark
NucleoBench enables cross-task comparisons under a shared protocol. Each algorithm receives the same 100 starting sequences per task, identical time budgets, and evaluations are repeated across five random seeds to track convergence speed and variability. The suite covers:
- Cell-type specific regulatory activity (Cell-type specific cis-regulatory activity).
- Transcription factor binding and chromatin accessibility.
- Selective gene expression using Enformer with inputs of about 200,000 base pairs and edit windows limited to 256 base pairs.
Beam search is a common strategy for constrained sequence editing. The team implemented both ordered and unordered variants, plus a hybrid approach called Gradient Evo.
Algorithm advances: AdaBeam
- Builds on unordered beam search with adaptive candidate selection and short, greedy exploration under a fixed compute budget.
- Integrates ideas from AdaLead, replacing its sampling step to achieve about 2x speed on long sequences.
- Uses gradient concatenation to reduce memory, enabling large-context models like Enformer.
Results at a glance
- AdaBeam outperformed existing methods on 11 of the 16 benchmark tasks.
- Comparators included simulated annealing, directed evolution, FastSeqProp, and Ledidi.
- Evaluations used identical starting sequences and fixed time budgets, tracking both convergence speed and variability across five seeds.
Why it matters
Designing DNA and RNA sequences with desired properties remains computationally challenging because the search space is enormous. For example, a 5' UTR region can exceed 2 x 10^120 possibilities. While predictive models have made great strides, standardized evaluation for design algorithms has been lacking. NucleoBench provides a controlled, cross-task yardstick, and AdaBeam shows practical gains in speed and scalability that could benefit applications from CRISPR gene therapies to mRNA vaccines.
Get the code and paper
- Announcement: Google Research blog
- Preprint: Paper
- Code: NucleoBench repository
- Package: AdaBeam design algorithm




.svg)